Pillar 2: Precision Medicine in Cardiology by Digital Twinning
Precision Medicine in Cardiology aims to account for individual variability in the pathophysiology of cardiovascular disease and is being enabled by the use of predictive patient-specific cardiac models [1]. However, current virtual heart tools are targeted to predict the optimal choice for a major, and often invasive, therapy and provide a snapshot model of the patient for facilitating decisions at a single time point only. Although Artificial Intelligence (AI) models built with large cohort studies, such as the UK Biobank, are providing valuable information on statistical relationships, these must be linked with computational models for precision medicine predictions on individuals. We propose to solve these problems by developing a cardiac digital twin [2]. A digital twin is a computer model that mimics a specific physical asset or entity and is updated with data from its physical twin to inform decisions that realise value. Healthcare is recognised as an industry likely to be disrupted by this technology [3], as it enables building continuously adjustable personalised and predictive models of patients based on tracked health and lifestyle data.
The team’s purpose is to build on their expertise in cardiac modelling [4], large cohort statistical modelling [5] and deep learning [6] to create a cardiac digital twin. Transitioning to the longitudinal scope of the digital twin is a critical step for longer term and continuous predictions, in order to match the patient over time as they age, and their disease is treated or develops.
The focus is on developing the data and modelling structures required to generate digital twins at scale, and applicable across several cardiovascular healthcare challenges. Importantly, emphasis will be given to mapping data to clinically explainable parameters, using physics-based models to solve the governing equations, thus avoiding the pitfalls of a data-driven “black box” digital heart twin approach. Current work includes:
- Develop innovative mechanistic patient evaluation workflows that enable digital twins to be updated with patient data over time and upscale the creation of cohorts of cardiac digital twins for clinical workflows.
- Use machine learning techniques to predict how the cardiac digital twin’s shape, tissue properties and boundary conditions progress due to disease, therapy, and ageing.
- Integrated workflow for creating posteriors from longitudinal patient images and diagnostic measurements and inferred model priors from patient demographics.
- Demonstration of cardiac digital twins and publication of publicly available virtual cohorts for heart failure (n=200), valve disease (n=200) and healthy (n=1000) cases.
The aim is to develop new methods for creating cardiac digital twins at scale, updating these digital twins over time and making long term predictions. The public virtual cohorts of healthy and failing heart anatomies and properties created in this extension will be an exceptional resource for academic and industry researchers. The generalisable methods developed for creating personalised models at the scale of clinical workflows will bring modelling and simulation applications closer to a clinical tool. The ability to update models and make long term predictions, using digital twins, will guide clinical timing decisions and predict both the amount and rate of response to therapy.
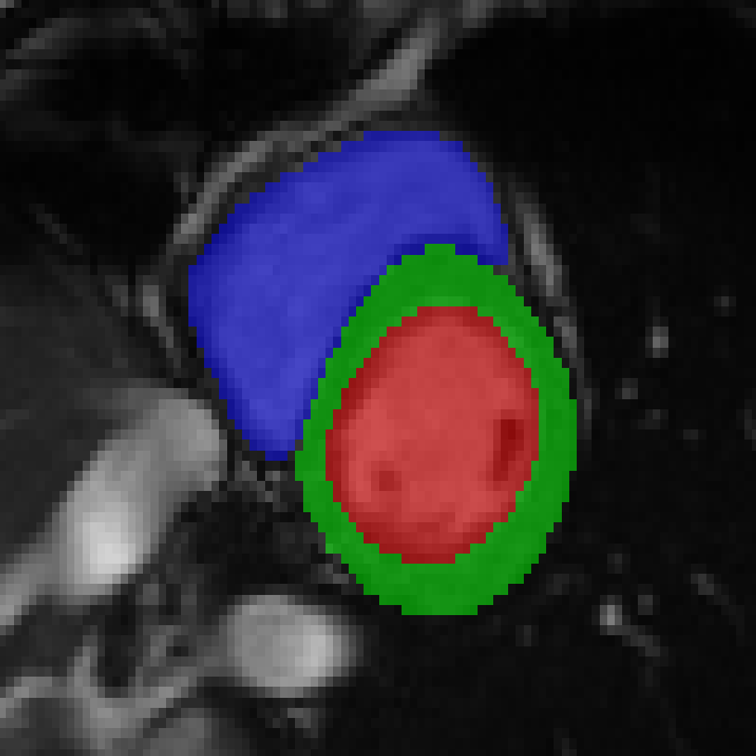
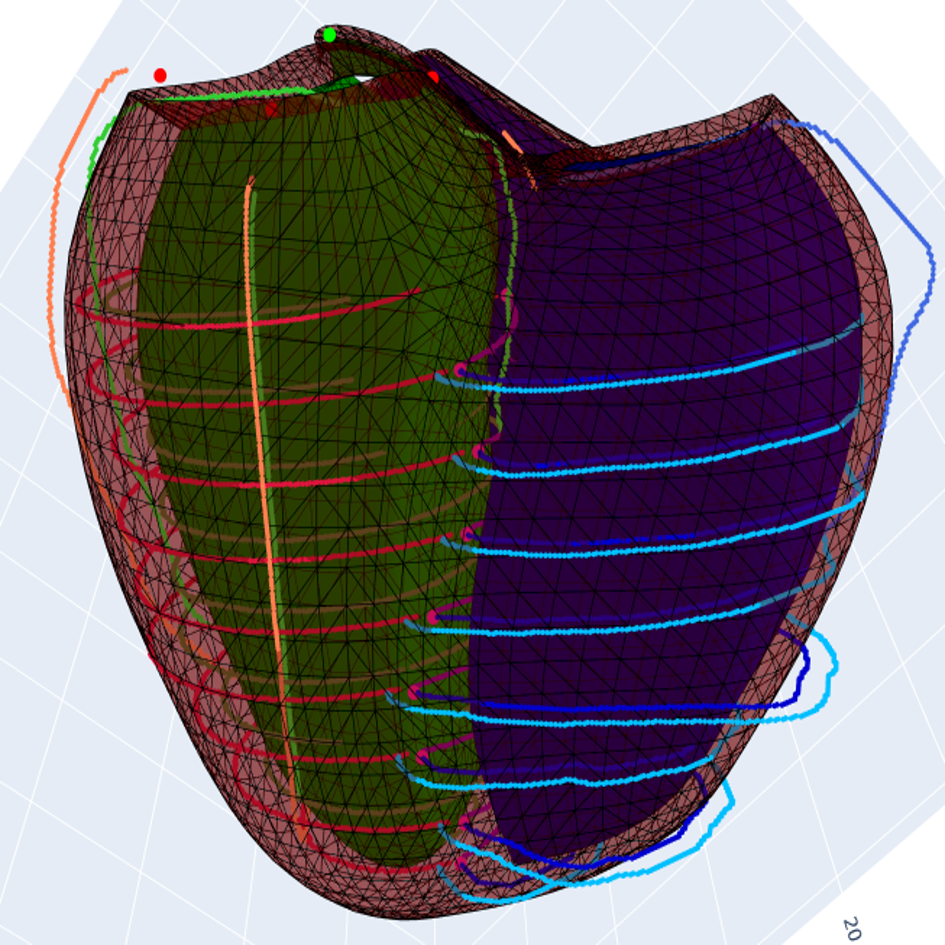
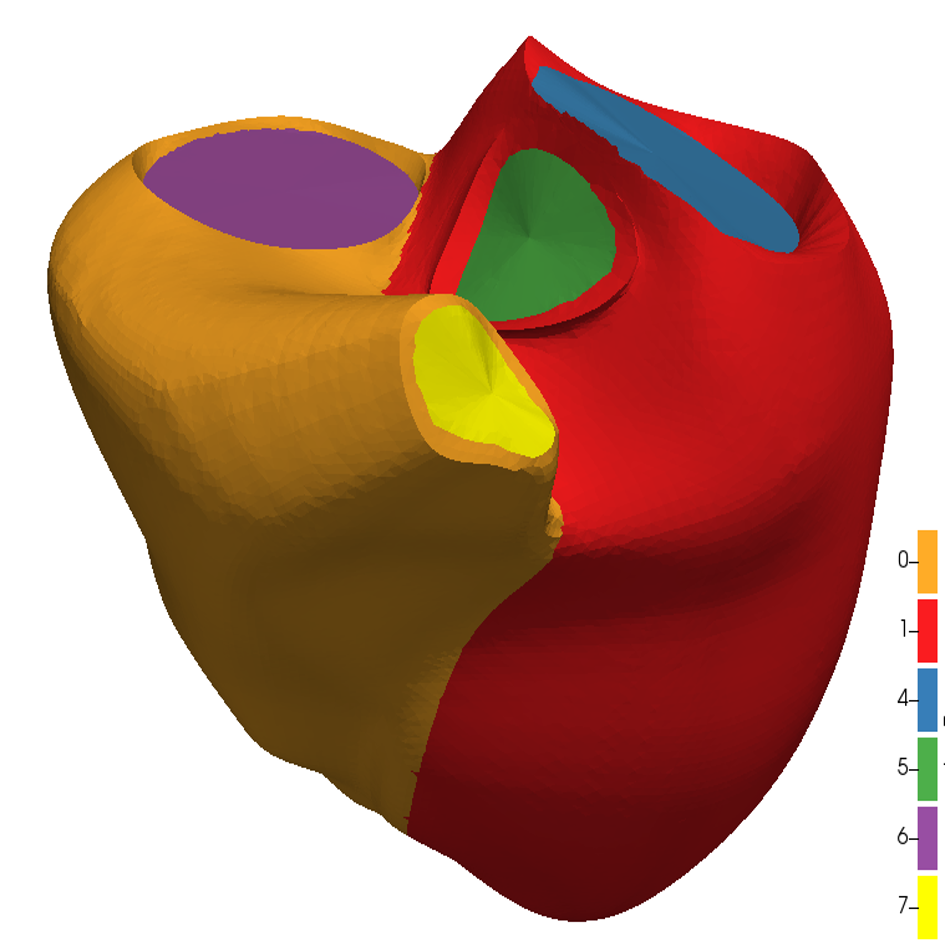
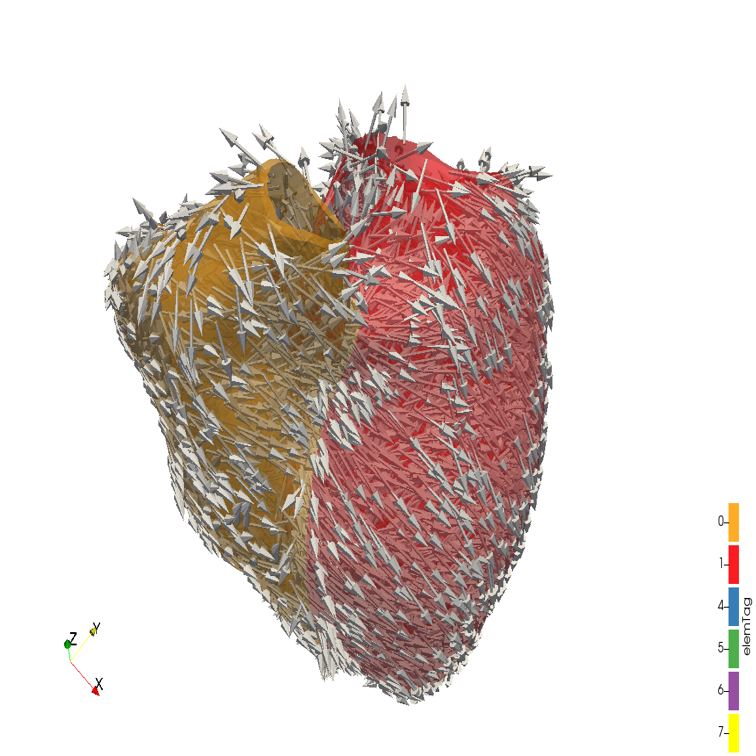
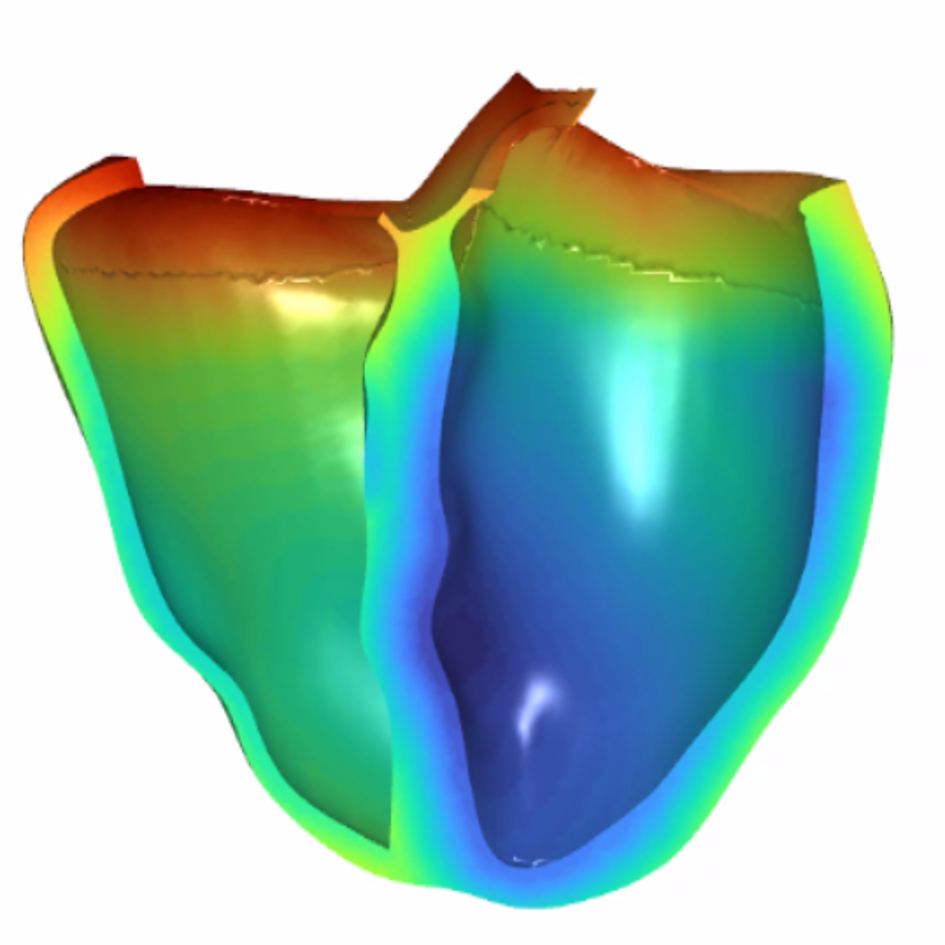
References
- Trayanova, N. (2019) ‘From genetics to smart watches: developments in precision cardiology’, Nature reviews. Cardiology, 16(2), pp. 72–73. doi: 10.1038/s41569-018-0149-y.
- Corral-Acero, J. et al. (2020) ‘The “Digital Twin” to enable the vision of precision cardiology’, European Heart Journal, 41(48), pp. 4556–4564. doi: 10.1093/eurheartj/ehaa159.
- Fuller, A. et al. (2020) ‘Digital Twin: Enabling Technologies, Challenges and Open Research’, IEEE Access, 8, pp. 108952–108971. doi: 10.1109/ACCESS.2020.2998358.
- Niederer, S. A., Lumens, J. and Trayanova, N. A. (2019) ‘Computational models in cardiology’, Nature Reviews. Cardiology, 16(2), pp. 100–111. doi: 10.1038/s41569-018-0104-y.
- Mauger, C. et al. (2019) ‘Right ventricular shape and function: cardiovascular magnetic resonance reference morphology and biventricular risk factor morphometrics in UK Biobank’, Journal of Cardiovascular Magnetic Resonance, 21. doi: 10.1186/s12968-019-0551-6.
- Puyol-Antón, E. et al. (2020) ‘Automated quantification of myocardial tissue characteristics from native T1 mapping using neural networks with uncertainty-based quality-control’, Journal of Cardiovascular Magnetic Resonance: Official Journal of the Society for Cardiovascular Magnetic Resonance, 22(1), p. 60. doi: 10.1186/s12968-020-00650-y.
